Cross-day or -session cell alignment alignment¶
The purpose of cross-session alignment is to allow users to align cells across imaging sessions (e.g. those taken on different days) and thus compare coding of cells, changing in biological variables, and other parameters across conditions and time in their data.
This page details additional notes on one of the algorithms used in CIAtah, usage outside the CIAtah GUI, and other tips. The main function used to run cross-session analysis that allows users to align cells across sessions/days can be found at:
Stable cell alignment across imaging sessions.
Below is an example of what the main output from cross-session alignment, a globalIDs matrix containing indices matches cells across days, looks like when visualized. Each column is an imaging session and each row is an individual global cell with the color indicating that global cell's within-session number. Any black cells indicate where no match was found for that global cell in that imaging day.
Algorithm overview¶
For details, see Cross-day analysis of BLA neuronal activity methods section in the Corder, Ahanonu, et al. Science, 2019:
We will also have details in an forthcoming imaging experiments and analysis book chapter.

Below is a general description of the algorithm/method used to match cells across sessions and references the above figure. A reference to the associated book chapter will be added in the future.
-
Load the cell extraction spatial filters and threshold them by setting to zero any values below 40% the maximum value for each spatial filter and use these thresholded filters to calculate each neuron's centroid location. You can calculate the centroid using the regionprops function in MATLAB. Do not round each neuron's centroid coordinates to the nearest pixel value as this would reduce accuracy of cross-day alignment.
-
Next, create simplified spatial filters that contained a 10-pixel-radius circle (this can be varied based on microns-per-pixel of the movie and size of cells) centered on each neuron's centroid location. This allows you to register different days while ignoring any slight day-to-day differences in the cell extraction algorithm's estimate of each neuron's shape even if the centroid locations are similar.
-
For each animal, we recommend that if you have N sessions to align that you choose the N/2 session (rounded down to the nearest whole number) to align to (referred to as the align session) to compensate for any drift or other imaging changes that may have occurred during the course of the imaging protocol.
-
For all imaging sessions create two neuron maps based on the thresholded spatial (see fig panel A, step 1, "thresholded neuron maps") and 10-pixel-radius circle (see fig panel A, step 2, "circle neuron maps") filters by taking a maximum projection across all x and y pixels and spatial filters (e.g. a max operation in the 3^rd^ dimension on a x × y × n neuron spatial filter matrix, where n = neuron number).
-
You then need to register these neuron maps to the align session using Turboreg with rotation enabled for all animals and isometric (projective) scaling enabled for a subset of animals in cases where that improves results. The registration steps are as follows (see fig panel A, step 3):
- Register the thresholded neuron map for a given session to the align session threshold neuron map.
- Use the output 2D spatial transformation coordinates to also register the circle neuron maps.
- Then register the circle neuron map with that animal's align session circle neuron map.
- Apply the resulting 2D spatial transformation coordinates to the thresholded neuron map.
- Repeat this procedure at least five times.
- Lastly, use the final registration coordinates to transform all spatial filters from that session so they matched the align session's spatial filters and repeat this process for all sessions for each animal individually.
-
After registering all sessions to the align session, re-calculate all the centroid locations (see fig panel A, step 4).
-
Set the align session centroids as the initial seed for all global cells (see fig panel A, step 5). Global cells are a tag to identify neurons that you match across imaging sessions.
- For example, global cell #1 might be associated with neurons that are at index number 1, 22, 300, 42, and 240 within the cell extraction analysis matrices across each of the first five imaging sessions, respectively.
-
Starting with the align session for an animal, calculate the pairwise Euclidean distance between all global cells' and the selected session's (likely 1^st^) neurons' centroids.
-
Then identify any cases in which a global cell is within 5 μm (nominally ~2 pixels in our data, this can be varied by the user) of a selected session's neurons. This distance depends on the density of cells in your imaging sessions, a stricter cut-off should be set for more dense brain areas. When you find a match, then check that the spatial filter is correlated (e.g. with 2-D correlation coefficient, Jaccard distance, or other measure) above a set threshold (e.g. r > 0.4) with all other neurons associated with that global cell (see fig panel A, step 6).
-
If a neuron passes the above criteria, add that neuron to that global cell's pool of neurons then recalculate the global cell's centroid as the mean location between all associated session neurons' centroid locations and annotate any unmatched neurons in that session as new candidate global cells.
-
Repeat this process for all sessions associated with a given animal.
Example output¶
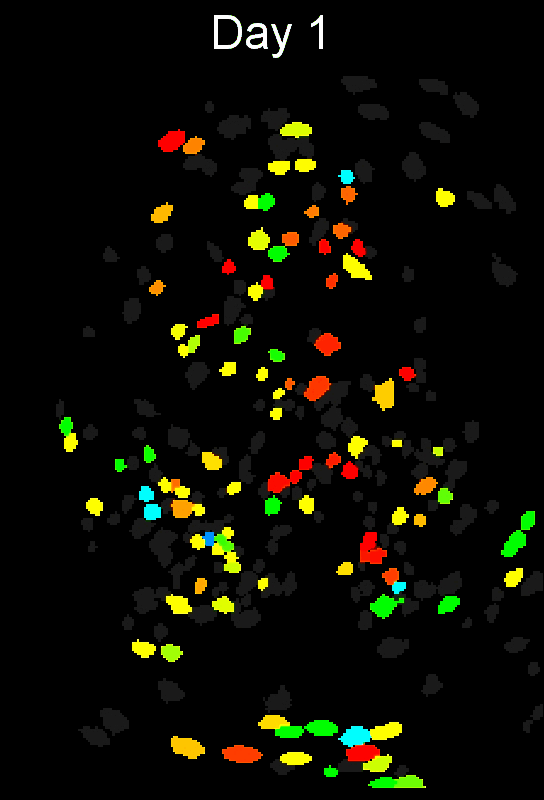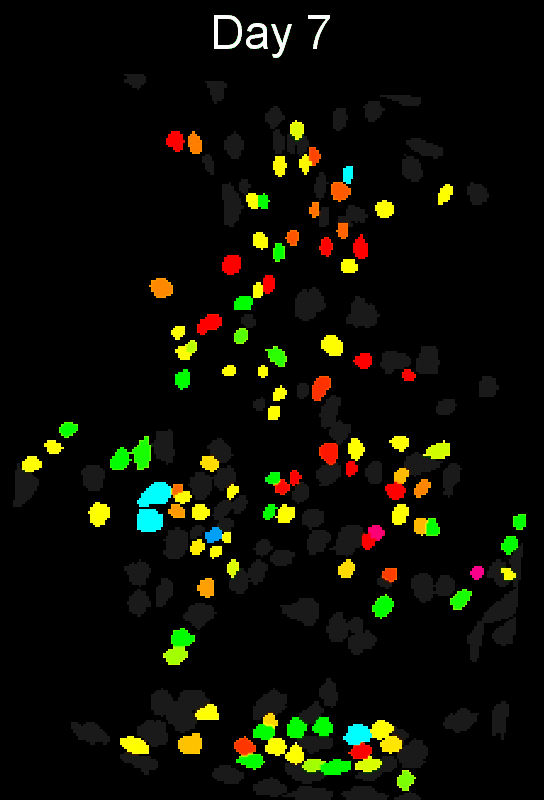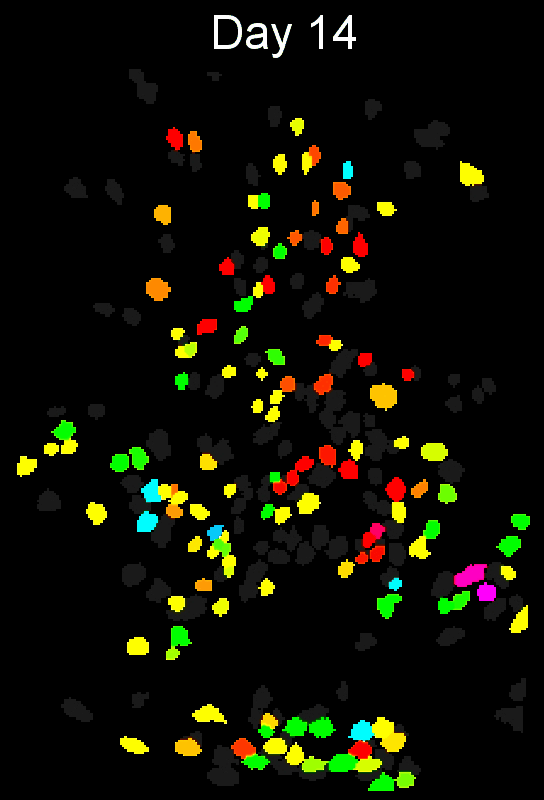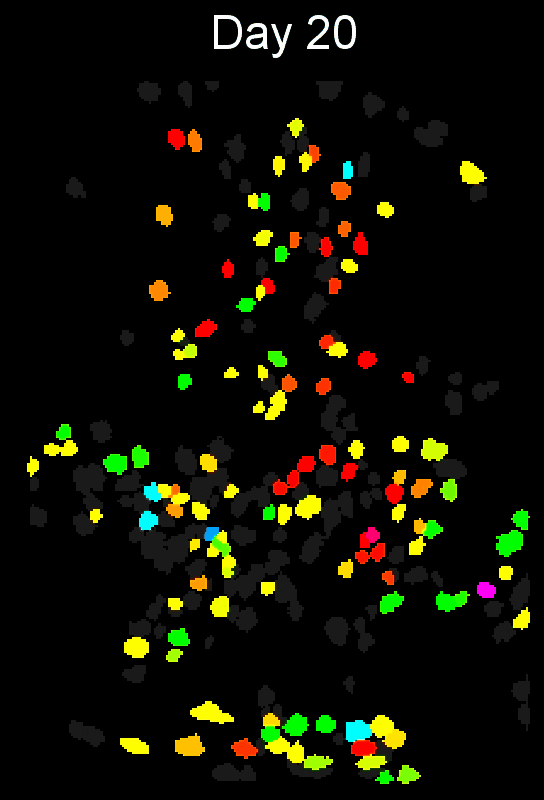
- Example output of several mPFC animals across multiple sessions. Color is used to indicate a global ID cell (e.g. the same cell matched across multiple days).

Usage¶
The below commands in MATLAB can be used to align sessions across days.
% Create input images, cell array of [x y nCells] matrices
inputImages = {day1Images,day2Images,day3Images};
% options to change
opts.maxDistance = 5; % distance in pixels between centroids for them to be grouped
opts.trialToAlign = 1; % which session to start alignment on
opts.nCorrections = 1; %number of rounds to register session cell maps.
opts.RegisTypeFinal = 2 % 3 = rotation/translation and iso scaling; 2 = rotation/translation, no iso scaling
% Run alignment code
[alignmentStruct] = matchObjBtwnTrials(inputImages,'options',opts);
% Global IDs is a matrix of [globalID sessionID]
% Each (globalID, sessionID) pair gives the within session ID for that particular global ID
globalIDs = alignmentStruct.globalIDs;
% View the cross-session matched cells, saved to `private\_tmpFiles` sub-folder.
[success] = createMatchObjBtwnTrialsMaps(inputImages,alignmentStruct);
In certain cases, you want to run analysis on the registered images, see below.
% Get registered images, cell array of [x y nCells] matrices
registeredImagesCell = alignmentStruct.inputImages;
% Get registered cell maps, cell array of [x y] matrices
registeredCellmaps = alignmentStruct.objectMapTurboreg;
% OR another method below.
% Get the registration coordinates
globalRegCoords = alignmentStruct.registrationCoords;
globalRegCoords = globalRegCoords{folderNo};
% Re-register input images for particular imaging session for later analysis.
for iterationNo = 1:length(globalRegCoords)
fn=fieldnames(globalRegCoords{iterationNo});
for i=1:length(fn)
localCoords = globalRegCoords{iterationNo}.(fn{i});
[inputImages, localCoords] = turboregMovie(inputImages,'precomputedRegistrationCooords',localCoords);
end
end
Algorithm results¶
Cross-session metrics and results on cross-session amygdala response to pain¶

PFC cross-session alignment.¶

Dorsal striatum cross-session algorithm comparison¶
- Original dorsal striatum cell maps from ICA with no motion correction applied.

CIAtah(Biafra's) registration algorithm- Color is used to indicate a global ID cell (e.g. the same cell matched across multiple days). Thus, same color cell = same cell across sessions under the quick iteration parameters used in the below run.

CellRegregistration algorithm Using older code at https://github.com/zivlab/CellReg.