CIAtah¶

Created by Biafra Ahanonu, PhD.
CIAtah (pronounced cheetah, formerly calciumImagingAnalysis [CIAPKG]) is a software package for analysis of one- and two-photon calcium imaging datasets.
Currently requires MATLAB and runs on all major operating systems (Windows, Linux [e.g. Ubuntu], and macOS).
Download the software at https://github.com/bahanonu/ciatah.

- Note:
CIAtahversionv4moves the remaining (i.e. all except external packages/software) CIAtah functions into theciapkgpackage to improve namespace handling and requires MATLAB R2019b or above (due to package import changes). Users with earlier versions of MATLAB can downloadCIAtahversionv3(see Releases) until pre-R2019b MATLAB support is fully integrated into v4.
- This user guide contains instructions to setup, run, and troubleshoot
CIAtah. - Note:
CIAtahis a class (ciatahwithin MATLAB) with various GUIs to allow processing of calcium imaging data. In addition, users can access the underlyingCIAtahfunctions to make custom workflows. See Custom command-line pipelines.
Guides¶
Below are recordings and additional documents for users who want to learn more about calcium imaging analysis/experiments and the CIAtah pipeline.
Book chapter — We have a book chapter that goes over all steps of miniscope imaging: viral injections, GRIN lens probe implant, pain experimental design, data processing and neural/behavioral analysis, and more.
- See Ahanonu, B., Corder, G. (2022). Recording Pain-Related Brain Activity in Behaving Animals Using Calcium Imaging and Miniature Microscopes (https://doi.org/10.1007/978-1-0716-2039-7_13).
Spinal cord motion correction methods — Methods for motion correction of spinal imaging data using feature identification (e.g. with DeepLabCut), control point registration, and other methods. Additional updates will be integrated into CIAtah in the future.
- Preprint: Ahanonu and Crowther, et al. (2023). Long-term optical imaging of the spinal cord in awake, behaving animals. bioRxiv (https://www.biorxiv.org/content/10.1101/2023.05.22.541477v1.full).
Webinars
-
My INSCOPIX INSPIRE webinar gives an overview of calcium imaging analysis (with a focus on CIAtah) along with tips for improving experiments and analysis: "Signal in the noise: Distinguishing relevant neural activity in calcium imaging".
-
Our INSCOPIX INSIGHTS seminar describing awake and freely moving spinal cord imaging: "Long-term, multicolor spinal cord neural imaging in freely moving animals".
Workshop tutorial — This recording gives an overview of setting up and using CIAtah: https://www.youtube.com/watch?v=I6abW3uuJJw.
Imaging analysis tools My table with many current imaging analysis tools: https://github.com/bahanonu/imaging_tools.
GRINjector — A surgical device to help with implanting gradient-refractive index (GRIN) lens probes into the brain or other regions: https://github.com/bahanonu/GRINjector.
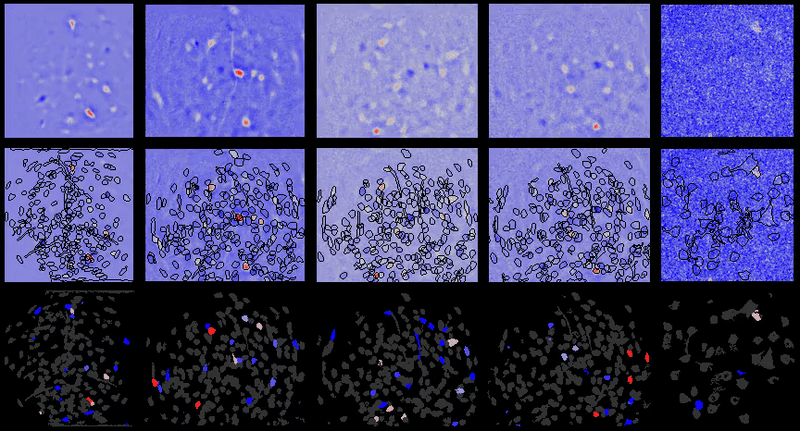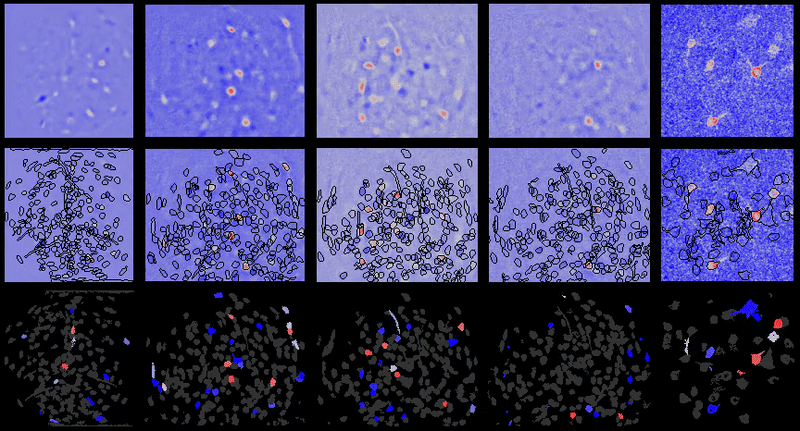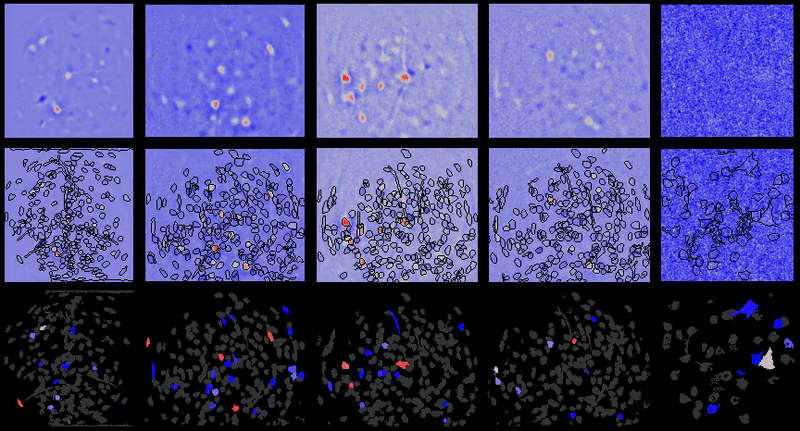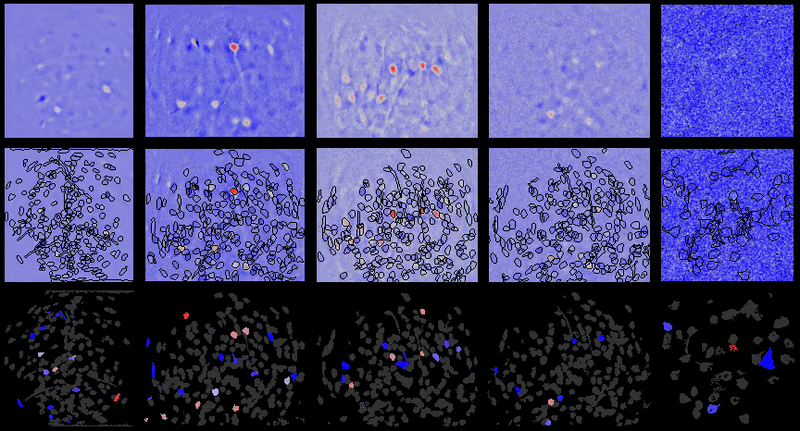
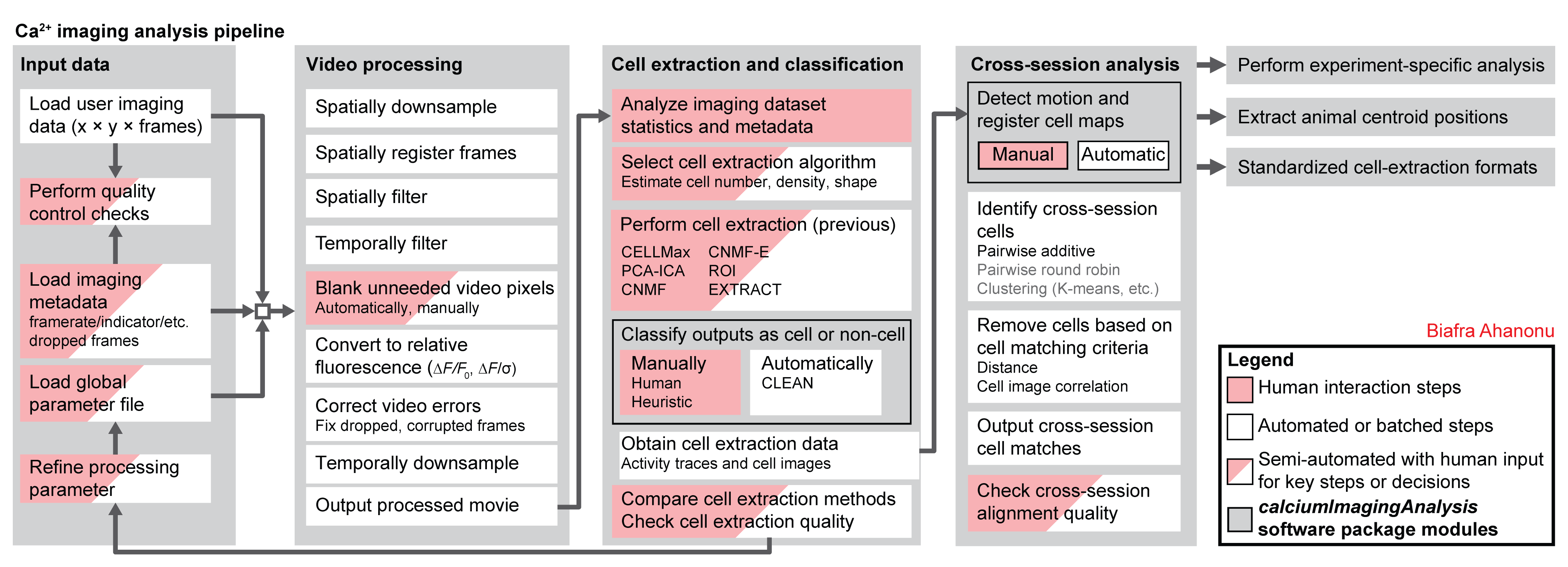
Movie processing, cell extraction, and analysis validation.
CIAtah cell sorting GUI
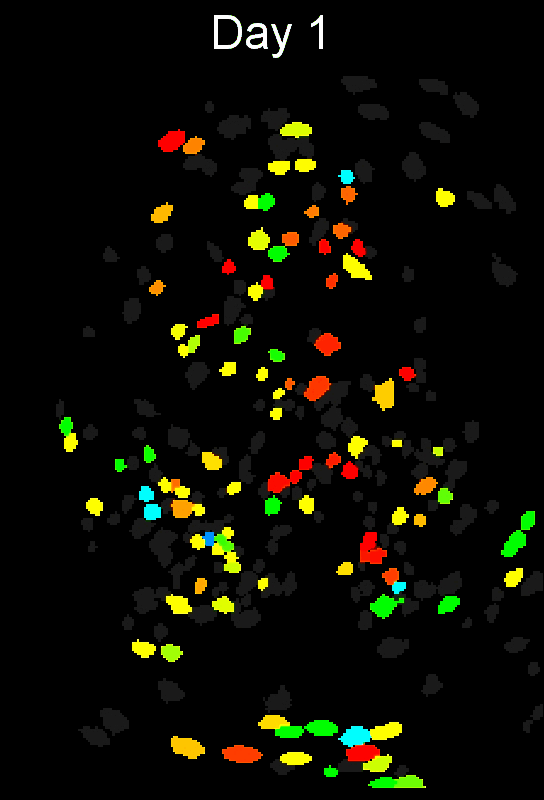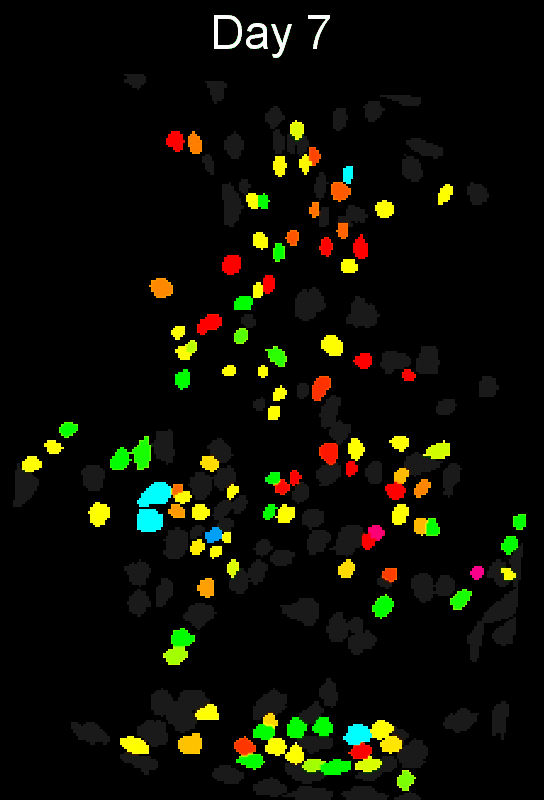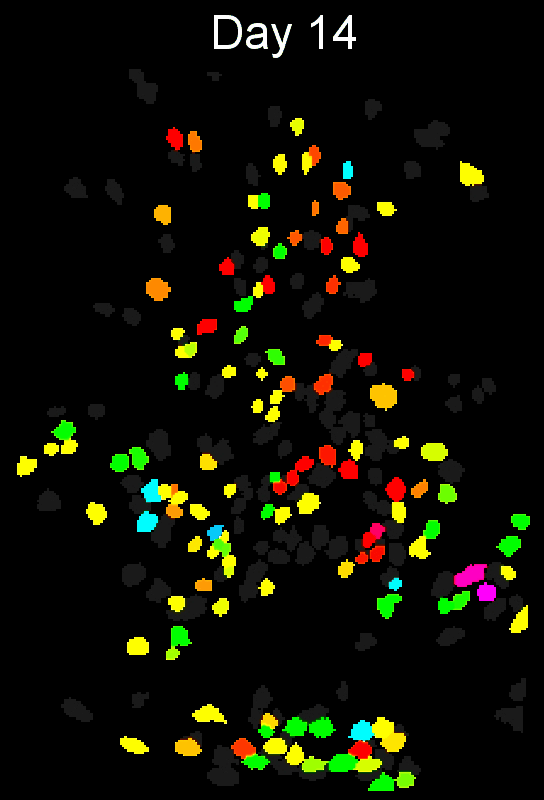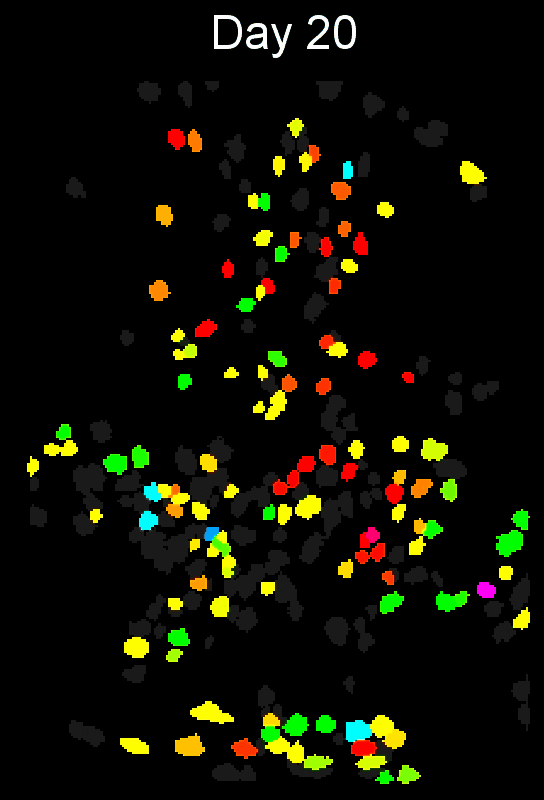
Stable cell alignment across imaging sessions.
CIAtah features:
- A GUI with different modules for large-scale batch analysis.
CIAtahfunctions (inciapkg/+ciapkgfolders) can be used to create GUI-less, command line-ready analysis pipelines.- Includes all major calcium imaging analysis steps:
- movie visualization (including reading from disk, for fast viewing of large movies)
- pre-processing (motion correction, spatiotemporal downsampling, spatial filtering, relative fluorescence calculation, etc.)
- support for multiple cell-extraction methods (CELLMax, PCA-ICA, CNMF, CNMF-E, EXTRACT, etc.)
- manual classification of cells via GUIs,
- automated cell classification (i.e. CLEAN algorithm, coming soon!),
- cross-session cell alignment, and more.
- Includes example one- and two-photon calcium imaging datasets for testing
ciatah. - Supports a plethora of major imaging movie file formats: HDF5, NWB, AVI, ISXD [Inscopix], TIFF, and Bio-Formats compatible formats (Olympus [OIR] and Zeiss [CZI and LSM] currently, additional support to be added or upon request).
- Supports Neurodata Without Borders data standard (see calcium imaging tutorial) for reading/writing cell-extraction and imaging movie files.
- Animal position tracking (e.g. in open-field assay).
- Requires
MATLAB.
Navigation¶
The main sections of the site:
Setup- installation ofCIAtah.Repository- notes about the software package and data formats.Processing data- sections related to processing calcium imaging movies using theCIAtahclass.API- details how to runCIAtahfrom the command line. Will include more details on the many underlying functions in the future.Help- several section that provide hints and help for processing calcium imaging.Misc- miscellaneous information about the repository.
References¶
Please cite our Corder, Ahanonu, et al. Science, 2019 publication if you used the software package or code from this repository to advance or help your research.
Questions?¶
Please open an issue on GitHub or email any additional questions not covered in the repository to bahanonu [at] alum.mit.edu.
Made in USA.

Repository stats¶
(starting 2020.09.16)
(starting 2020.09.22)
(starting 2023.06.12, specific to docs homepage)